CRISPR-CAS9-MEDIATED GENOME EDITING
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and CRISPR-associated (Cas) genes are required for bacterial and archeal innate immunity against foreign DNA (Marraffini, Nature, 2015). During the acquisition phase, foreign DNA fragments are incorporated into CRISPR loci located in the bacterial and archeal genome. Transcribed RNA from CRISPR loci is subsequently processed into CRISPR RNAs, which are then incorporated into Cas proteins. The most studied Cas protein is Cas9, a DNA endonuclease that forms a complex with CRISPR RNAs to cleave foreing DNA at specific sequences recognized by CRISPR RNAs (Figure 11) (Jinek et al, Science, 2012). CRISPR-Cas9 has been engineered to introduce sequence specific DNA double-strand breaks (DSBs) in all cellular and animal models currently studied, therefore allowing site-specific genome editing by homology-directed repair (HDR) or non-homologous end joining (NHEJ) (Wright et al, Cell, 2016; Hsu et al, Cell, 2014).

Figure 11. CRISPR-Cas9-mediated DNA double-strand break formation and repair
INACTIVATION OF EUKARYOTIC GENES BY INDUCTION OF STOP CODONS (iSTOP)
As an alternative to HDR- and NHEJ-dependent genome editing, CRISPR-dependent editing strategies that entail direct modification of DNA bases have recently been developed (Hess et al, Nature Methods, 2016; Komor et al, Nature, 2016; Ma et al, Nature Methods, 2016; Nishida et al, Science, 2016; Yang et al, Nature Comm, 2016). Distinct from standard CRISPR-Cas9-dependent genome editing, CRISPR-mediated base editing avoids the formation of DSBs, thus resulting in reduced genomic rearrangements and cell death (Kuscu et al, Nature Methods, 2017). CRISPR-dependent base editors consist of a catalytically inactive form of Cas9 or a Cas9 nickase mutant fused to cytidine deaminases, such as APOBEC1 or AID. In particular, the CRISPR-dependent base editor BE3 is a fusion of rat APOBEC1 (rAPOBEC1), a uracil glycosylase inhibitor (UGI) and the Cas9-D10A nickase mutant (Komor et al, Nature, 2016; Figure 12). Our studies have shown that CRISPR-dependent base editing efficiently inactivates genes by precisely converting four codons (CAA, CAG, CGA and TGG) into STOP codons without DSB formation (Billon, Bryant et al, Mol Cell, 2017; Figure 12). To facilitate gene inactivation by induction of STOP codons (iSTOP), we have generated a database of over 3.4 million sgRNAs for iSTOP (sgSTOPs) targeting 97-99% of genes in 8 eukaryotic species (https://www.ciccialab-database.com/istop). This database includes annotations for off-target propensity, percentage of isoforms targeted, prediction of nonsense-mediated decay and restriction enzymes that allow the rapid detection of iSTOP-mediated editing in cell populations and clones. Additionally, our database includes sgSTOPs that could be employed to precisely model over 32,000 cancer-associated nonsense mutations. This work provides a comprehensive resource for DSB-free gene disruption by iSTOP.
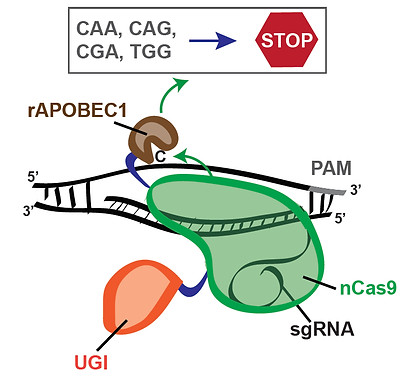
Figure 12. Induction of STOP codons by CRISPR-mediated base editing
DETECTION OF MARKER-FREE PRECISION GENOME EDITING AND GENETIC VARIATION USING DTECT
Detecting precise genomic modifications often requires sophisticated, expensive, and time-consuming experimental approaches. To overcome these limitations, we developed DTECT (Dinucleotide signaTurE CapTure), a rapid and versatile detection method that relies on the capture of targeted dinucleotide signatures resulting from the digestion of genomic DNA amplicons by the type IIS restriction enzyme AcuI (Billon et al, Cell Reports, 2020, Figure 13). DTECT enables the accurate quantification of marker-free precision genome editing events introduced by CRISPR-dependent homology-directed repair, base editing, or prime editing in various biological systems, such as mammalian cell lines, organoids, and tissues. Furthermore, DTECT allows the identification of oncogenic mutations in cancer mouse models, patient-derived xenografts, and human cancer patient samples. The ease, speed, and cost efficiency by which DTECT identifies genomic signatures should facilitate the generation of marker-free cellular and animal models of human disease and expedite the detection of human pathogenic variants.
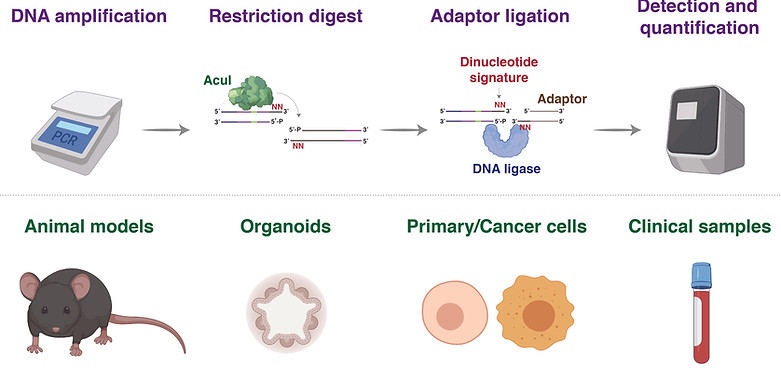
Figure 13. Identification and quantification of nucleotide variants in the indicated biological systems using DTECT
CURRENT STUDIES
Definition of cellular pathways that regulate CRISPR-mediated gene editing
We are currently employing genetic and biochemical tools to define the cellular machineries that repair DSBs formed using CRISPR-Cas9 technology and identify factors that determine whether Cas9-induced DSBs are repaired by HDR or NHEJ. These studies will provide novel insights into CRISPR-mediated genome editing and lead to the development of improved CRISPR-based technologies.
In initial studies, we individually expressed in human cells 204 open reading frames involved in the DDR and determined their impact on CRISPR-mediated HDR. From this work, we identified RAD18 as a stimulator of CRISPR-mediated HDR (Nambiar et al, Nature Comm, 2019; Figure 14). By defining the RAD18 domains required to promote HDR, we derived an enhanced RAD18 variant (e18) that stimulates CRISPR-mediated HDR in multiple human cell types, including embryonic stem cells. Mechanistically, e18 induces HDR by suppressing the localization of the NHEJ-promoting factor 53BP1 to DSBs. This study identified e18 as an enhancer of CRISPR-mediated HDR and highlighted the promise of engineering DDR factors to augment the efficiency of precision genome editing.

Figure 14. DDR ORF screen identifies RAD18 as one of the top HDR enhancers
Large-scale analysis of DDR nucleotide variants using base editing screens
We have recently developed CRISPR-dependent base editing screening technologies to study the function of nucleotide variants in human genes (Cuella-Martin et al, Cell, 2021). In these studies, we introduced thousands of nucleotide variants in 86 DDR genes and performed large-scale phenotypic analyses of DDR mutational outcomes upon treatment with DNA damaging agents commonly used for cancer therapy. These screens allowed us to identify mutations with loss-, gain- and separation-of-function (LOF, GOF, SOF) patterns, define novel protein domains and identify pathogenic variants of previous unknown clinical significance (Figure 15). We are now investigating the function of the identified DDR variants using genetic and cell biological approaches to define their impact on human disease.

Figure 15. Identification of new protein domains, LOF, GOF and SOF mutations, and pathogenic variants using base editing screens
